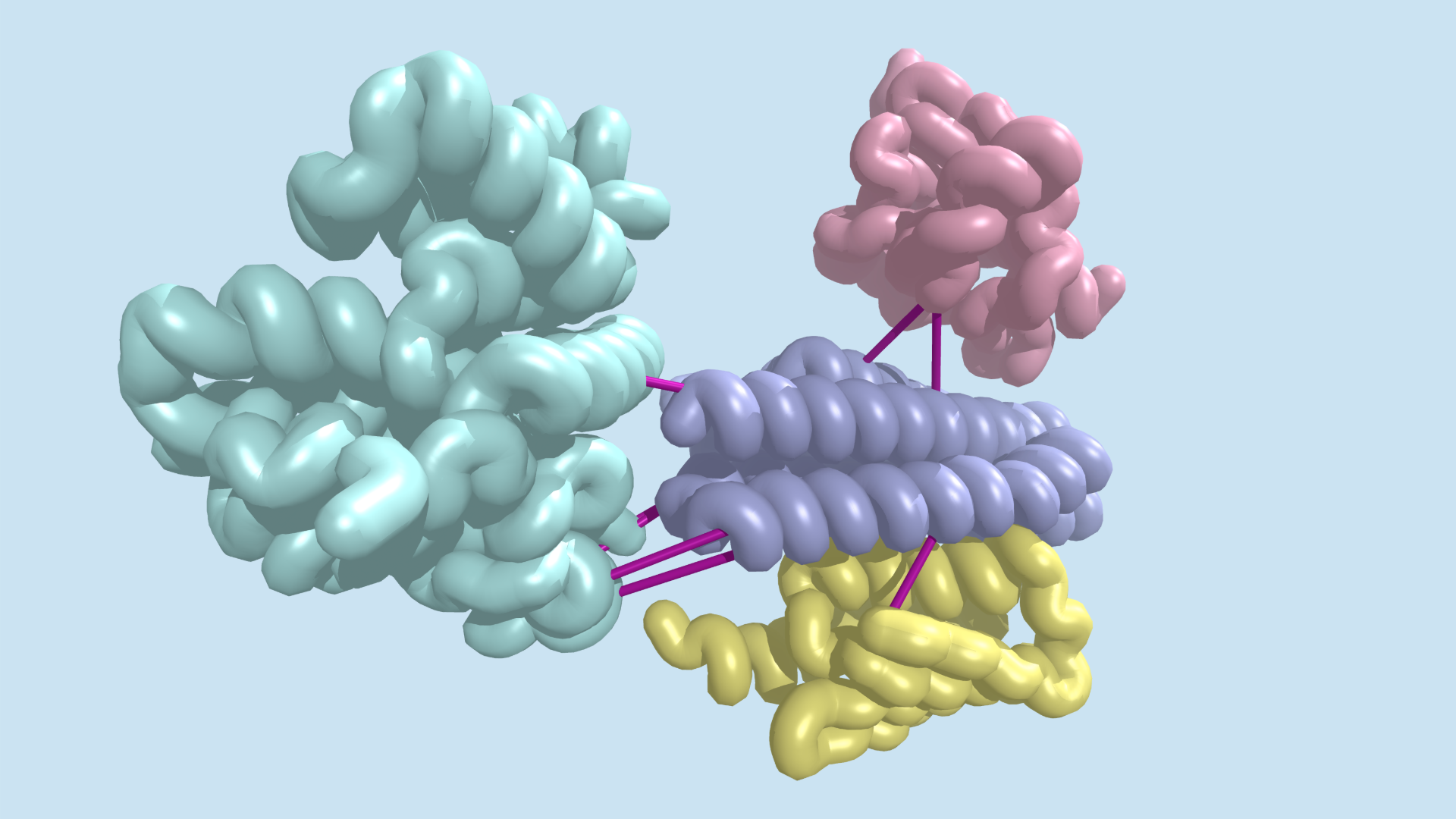
Coevolution at the proteome scale - Institute for Protein Design
Price: $ 43.00
4.5(556)
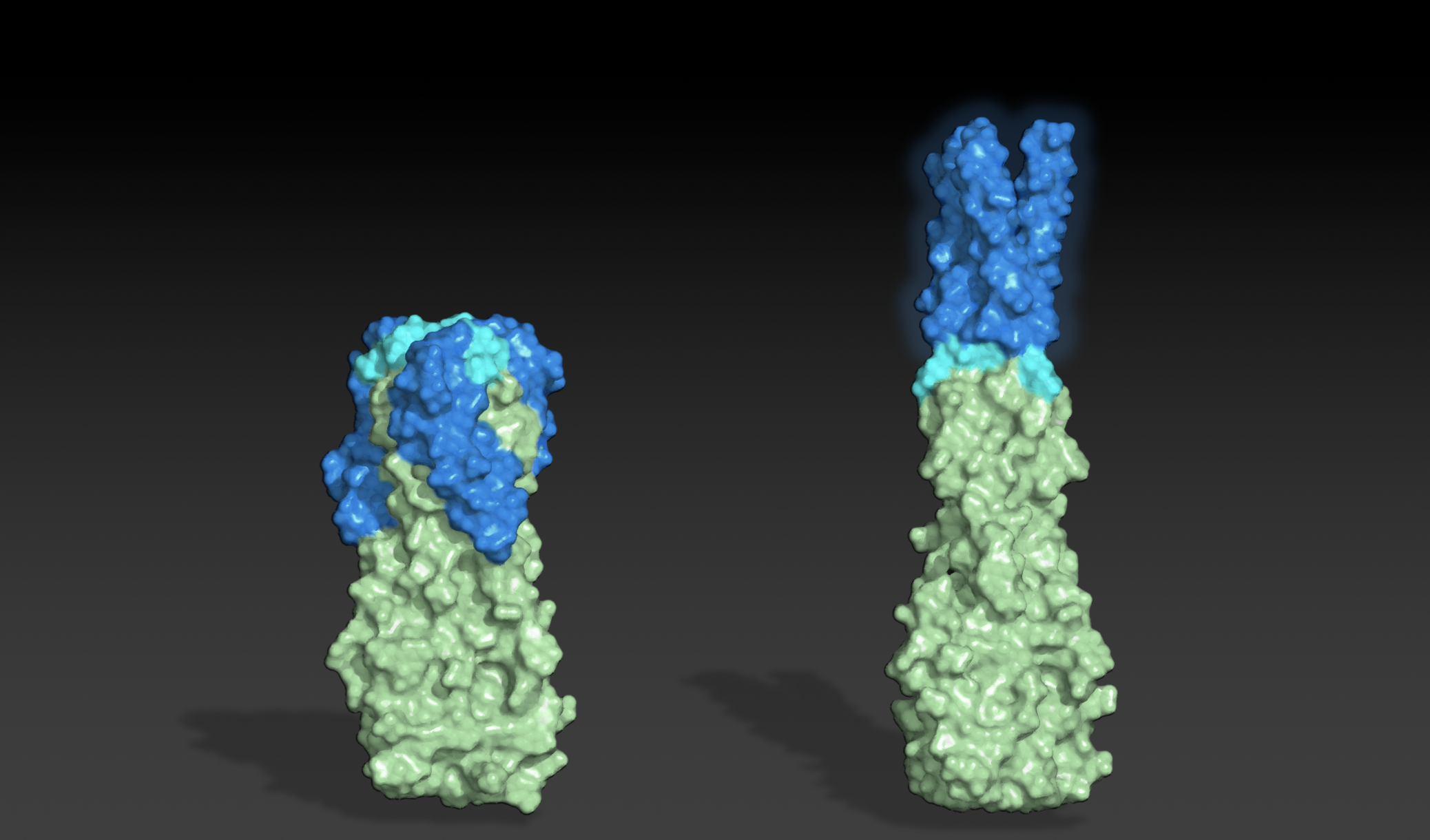
Today we report in Science the identification of hundreds of previously uncharacterized protein–protein interactions in E. coli and the pathogenic bacterium M. tuberculosis. These include both previously unknown protein complexes and previously uncharacterized components of known complexes. This research was led by postdoctoral fellow Qian Cong and included former Baker lab graduate student Sergey Ovchinnikov, now a John Harvard Distinguished Science
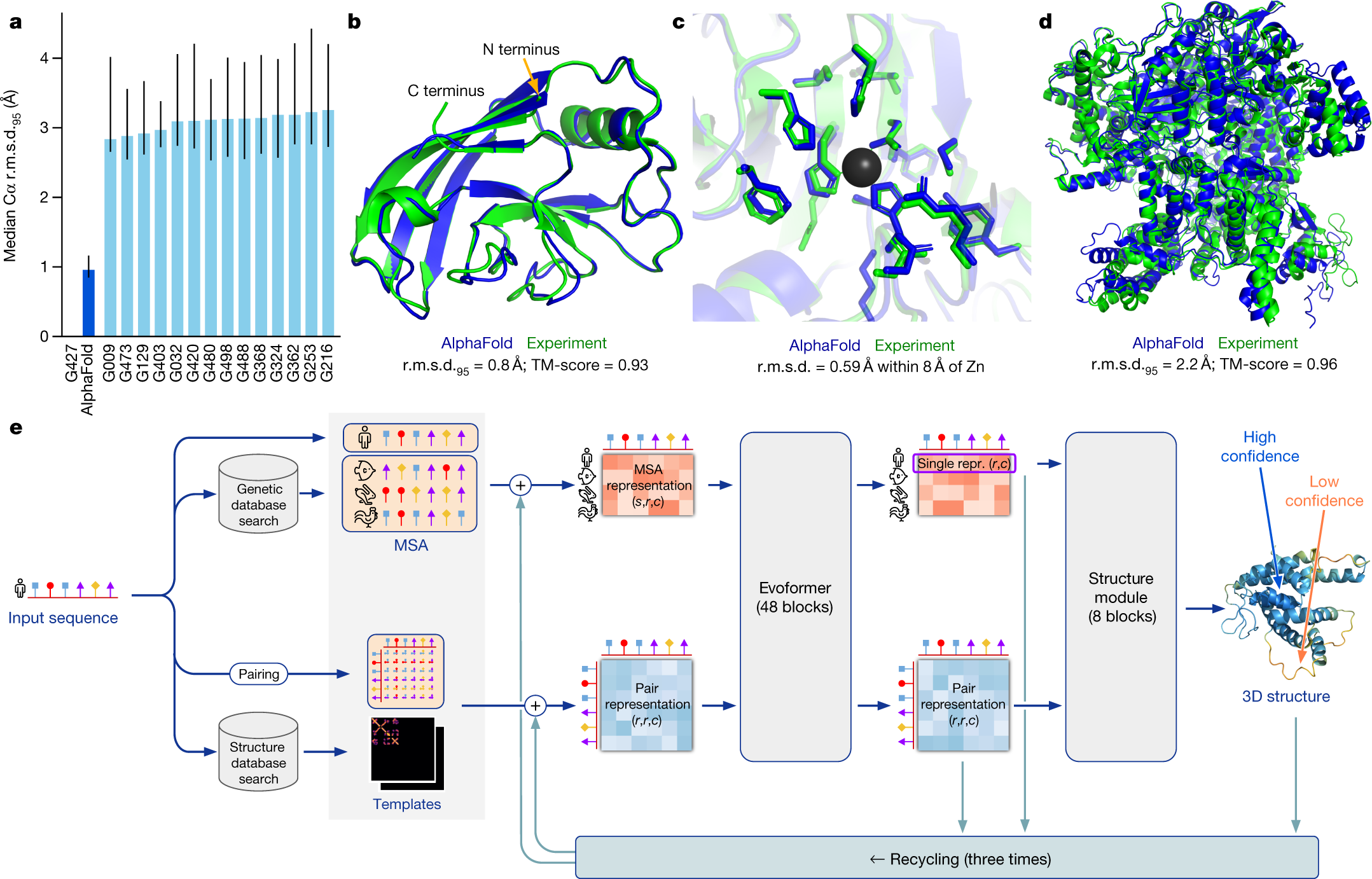
Highly accurate protein structure prediction with AlphaFold

Deploying synthetic coevolution and machine learning to engineer protein- protein interactions

Deploying synthetic coevolution and machine learning to engineer protein- protein interactions

Recent advances in predicting and modeling protein–protein interactions: Trends in Biochemical Sciences

Computed structures of core eukaryotic protein complexes

IPD Publication - Institute for Protein Design
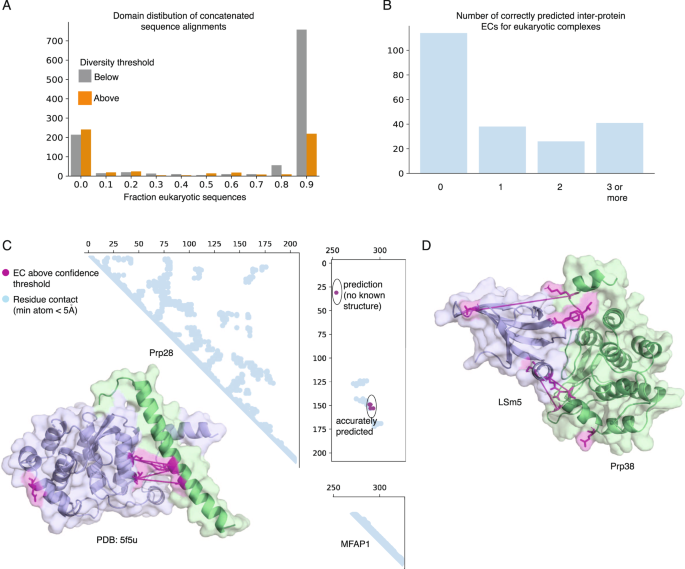
Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences
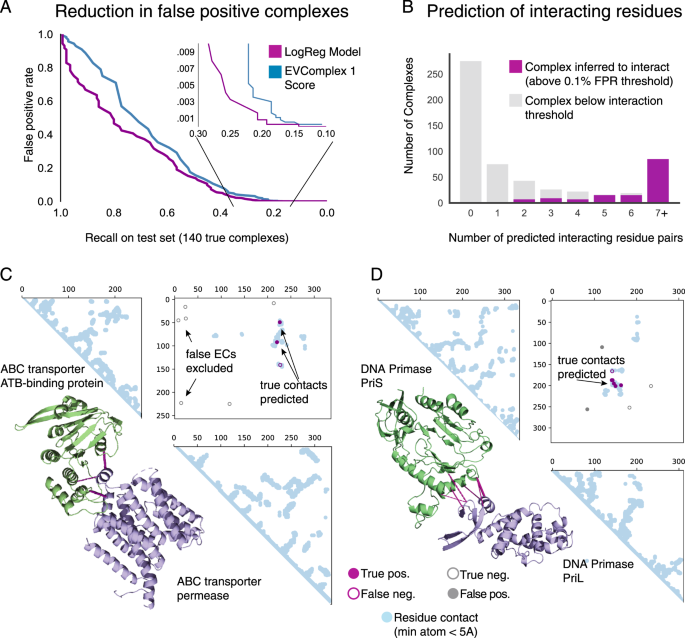
Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences

Coevolution at the proteome scale - Institute for Protein Design

Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences
News Roundup – Baker Lab

Computed structures of core eukaryotic protein complexes

PDF) Emerging Methods in Protein Co-Evolution




